· Search
· Download
· Views
· Homologs
Overview
The genome browser contains 3 P. pastoris genomes:
· Komagataelle phaffii CBS7435
· Komagataella phaffii GS115
· Komagatella pastoris DSMZ 70382
And 1 Saccharomyces cerevisea genome SC 288C.
For every genome different data tracks, containing diverse information, are available. You can select the tracks on the menu on the left side.
How
to display/select a strain or species in the Stand-Alone JBrowse
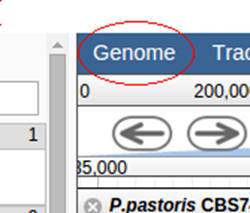
Click „Genome“ in the top menu –
select the desired strain or species.
The left menu will change according the available information for the strain or species.
How To Search for a feature
There are several options to search for features like genes etc.
1.
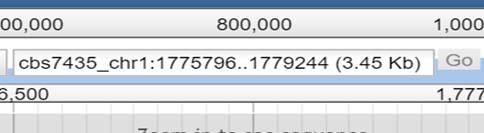
Enter the combination
of chromosome/contig/scaffold plus the region range you want
to view. Example: cbs7435:0..100
It is also possible to select
the contig/chromosome/scaffold from the drop down menu on the left side of the textbox you can
see on the right side.
2.
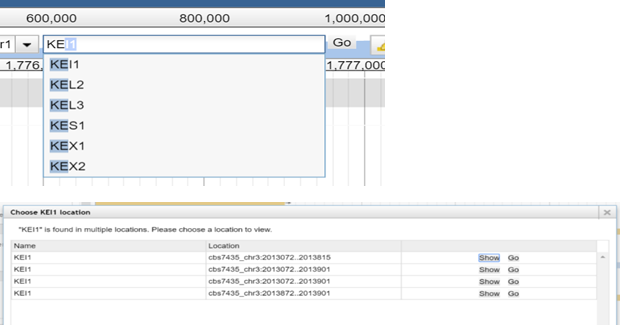
Enter the gene name of the feature you are looking
for into the textbox. While typing you will see matching suggstions.
You can also type in the feature name.
Select the match from the drop down list and click „Go“. A new PopUp window will open. There you can
choose between „Show“ and
„Go“ for the found features matching your searchterm. „Go“ will bring you directly to
the chosen feature and close the pop up,
„Show“ will do the same while leaving
the pop up open.
How to Retrieve/Download Sequences
1.
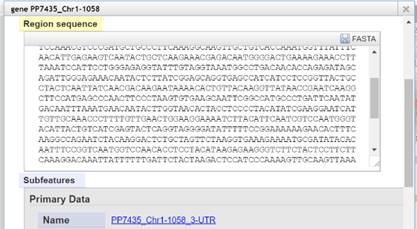
In „View Details“
Right Mouse Click on the feature
you want the sequence for. Choose
„View Details“ – a window opens
containing one or more versions
of the feature sequence where you can
do a copy and paste. If there is
no context menu „View Details“ by right clicking a track, you can
do a simple left mouse click on the feature to open the details window.
2.
In GFF-, BED-, Sequin-Format
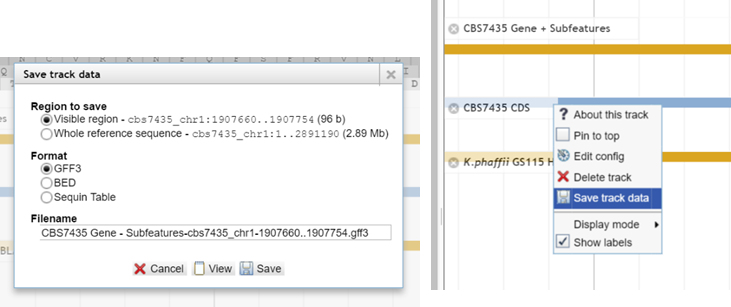
Open the context menu of some track – not reference sequence track - and choose „Save track data“. You
have the option to save the sequence in the three formats. Also you can choose
if you want
to save only the visible region or the complete reference sequence.
3.
Whole Reference Genome
See the paragraph above.
4.
Nothing above
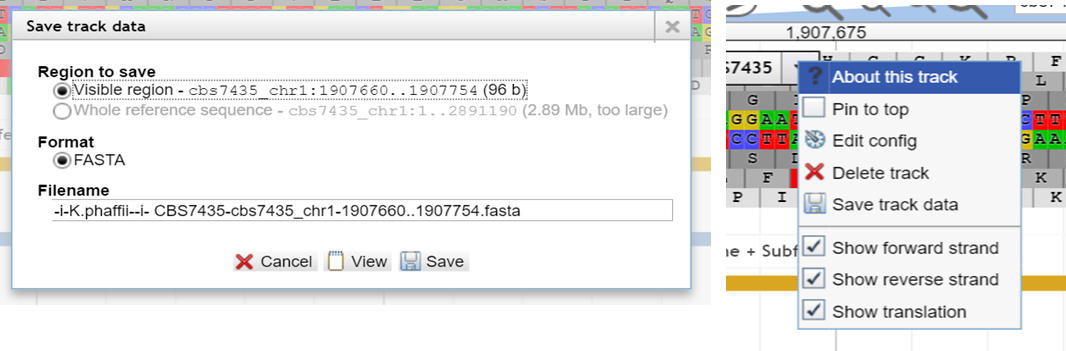
If you need
a chunk of sequence which is not part
of a feature-track, you can fetch it
by utilizing the reference sequence. Zoom into the exact section you need,
then select the reference sequence track on the left menu. Now you
can download thesequence in fasta format by clicking
the drop down menu, choose "Save track data" and clicking
"View" or "Save"
How to navigate
in JBrowse
1.
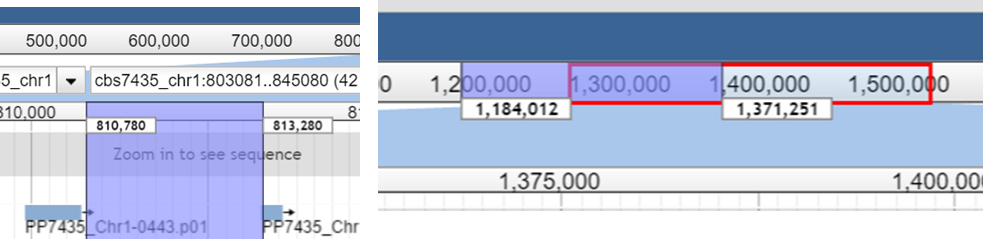
Zoom in and out
- Double Click multiple times at the position you want
to zoom in
- Magnifier button: Press
+/- on the magnifier buttons
- Drag & Drop the coursor either
on the upper or the lower coordinate ruler
2.
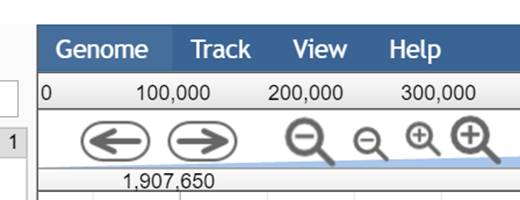
Moving left/right
- Click the arrows to the left or right
- Drag & Drop the page at empty
position with the coursor
3.
Hiding track menu to
prevent it overlapping data, use the „Hide“-Button
![]()
Views
1. Different Tracks have a different appearance depending on the type of data and the zoom status
2. A details view can be reached by right clicking on the track, apart from the „mRNA + SeqLighter“ Tracks. In the „mRNA + SeqLighter“ just use the left mouse button instead.
3. In the „mRNA + SeqLighter“ track you can reach the SeqLighter-Plugin via the right-click context menu – choose „View Sequence“. Beside the possibility to download the sequence in different formats you can choose some ranges on flanking regions tob e displayed and downloaded as well.
Sharing and Marking information
You can send a link to the current view you are looking at using the „Share“-Button on the right upper corner.
![]()
You can mark
a region using the highlighter-button next to the search textbox

Data Sources
1. Pichia pastoris
CBS7435
The CBS7435 strain was sequenced
by the Institute for Genomics and Bioinformatics, Graz
University of Technology (Austria).
Reference: High-quality
genome sequence of Pichia pastoris
CBS7435, Journal of Biotechnology 2011, 154(4):312-20
The hosted sequence
version is from 27-FEB-2015 (NCBI)
The annotation was curated 2016. This data is part of this database and genome browser.
Reference: Valli et.al.,Curation of the genome annotation of Pichia pastoris (Komagataella phaffii) CBS7435 from gene level to protein function
2. Pichia pastoris
GS115
The GS115 strain was sequenced
by the VIB Ghent, Department
of Biomedical Molecular Biology.
Reference: De
Schutter et al.,Genome
sequence of the recombinant
protein production host Pichia pastoris,
Nature Biotechnology 27, 561 - 566 (2009)
The hosted sequence
version is from 27-FEB-2015 (NCBI)
3. Pichia pastoris
DSMZ70382
The DSMZ 70382 strain was sequenced
by the University of Natural Resources and Applied
Life Sciences, Vienna, Department of Applied Microbiology.
Reference: Mattanovich et al., Genome, secretome
and glucose transport highlight unique features of the protein production host Pichia pastoris, Microbial Cell Factories 2009, 8:29
(NCBI)
4. Saccharomyces cerevisiae
S288c
The sequenceinformation of S.cerevisiae S288c was downloaded
from yeastgenome.org (genome, Coding Sequences) on February, 10th 2016
Homologs in other Strains & Species
The homologs were derived from a simple BLAT run of the according coding sequences (CDS) against a genomic sequence (all in nucleotide sequences). Blast was chosen as output format, so that a previously used Blast parser could be used to determine the hits.
Example call:
blat cbs7435_genomic.fasta sc_orf_coding_all.fasta -t=dna -q=dna -out=blast sc_cds_vs_cbs7435.blast
The result was parsed the way, that only the best HSPs got into the output. The used cut offs were:
· Fraction conserved aka fraction similar of 0.4
· Length of 50
Software Sources
1. JBrowse
2. SeqLighter: A plugin developed by Maria Kim and Vivek Krishnakumar for the Araport Website (URL)